library(ggplot2)해당 강의노트는 전북대학교 이영미교수님 2022-2 고급회귀분석론 자료임
import
데이터
############ 데이터 불러오기
dt <- data.frame(x = c(4,8,9,8,8,12,6,10,6,9),
y = c(9,20,22,15,17,30,18,25,10,20))
dt| x | y |
|---|---|
| <dbl> | <dbl> |
| 4 | 9 |
| 8 | 20 |
| 9 | 22 |
| 8 | 15 |
| 8 | 17 |
| 12 | 30 |
| 6 | 18 |
| 10 | 25 |
| 6 | 10 |
| 9 | 20 |
cor(dt$x, dt$y)
0.921812343945765
plot(y~x,
data = dt,
xlab = "광고료",
ylab = "총판매액",
pch = 16,
cex = 2,
col = "darkorange")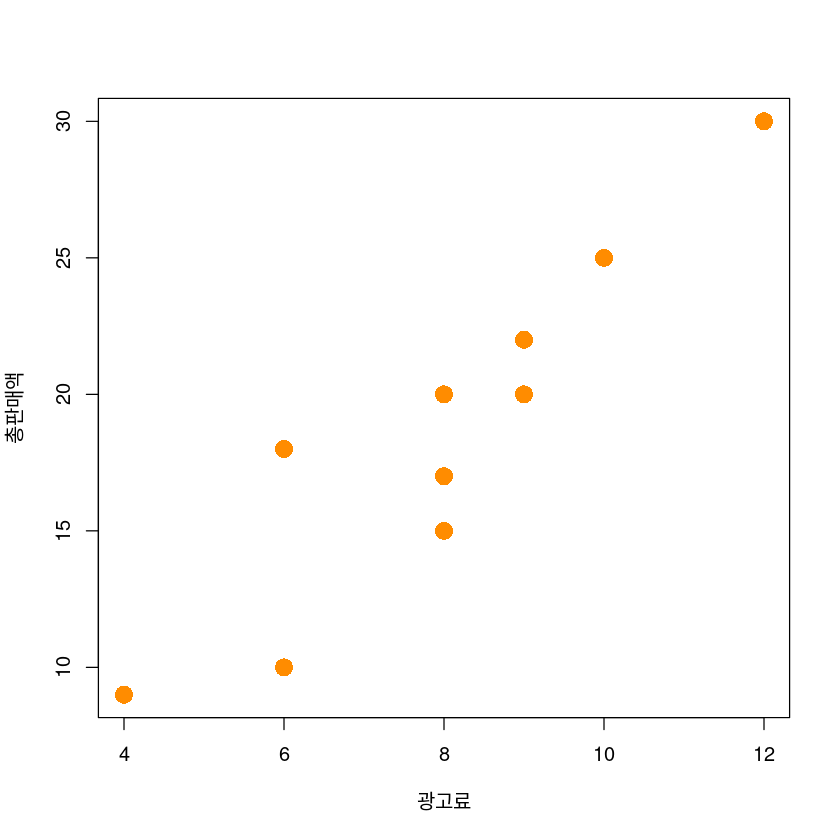
ggplot(dt, aes(x, y)) +
geom_point(col='steelblue', lwd=2) +
# geom_abline(intercept = co[1], slope = co[2], col='darkorange', lwd=1.2) +
xlab("광고료")+ylab("총판매액")+
# scale_x_continuous(breaks = seq(1,10))+
theme_bw() +
theme(axis.title = element_text(size = 14))Warning message in geom_point(col = "steelblue", lwd = 2):
“Ignoring unknown parameters: `linewidth`”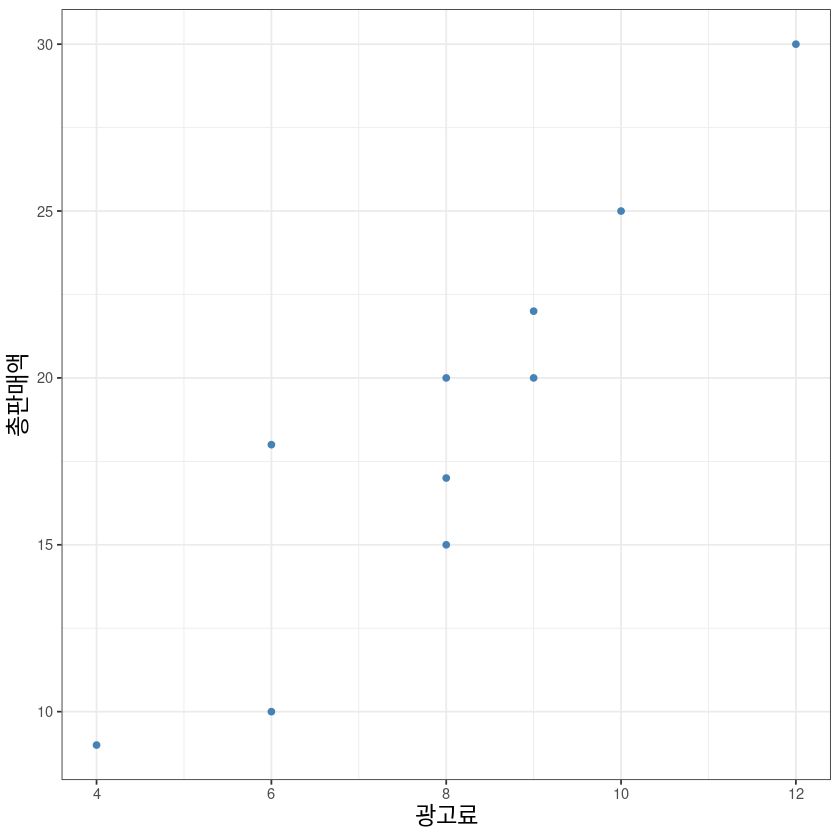
적합
hat y = hat (E(y|X=x)) = hat beta_0 +hat beta_1 * x
H0 : beta0 =0 vs H1 : beta0 != 0
H0 : beta1 =0 vs H1 : beta1 != 0
## y = beta0 + beta1*x + epsilon
model1 <- lm(y ~ x, dt)
model1
Call:
lm(formula = y ~ x, data = dt)
Coefficients:
(Intercept) x
-2.270 2.609 summary(model1)
Call:
lm(formula = y ~ x, data = dt)
Residuals:
Min 1Q Median 3Q Max
-3.600 -1.502 0.813 1.128 4.617
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -2.2696 3.2123 -0.707 0.499926
x 2.6087 0.3878 6.726 0.000149 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 2.631 on 8 degrees of freedom
Multiple R-squared: 0.8497, Adjusted R-squared: 0.831
F-statistic: 45.24 on 1 and 8 DF, p-value: 0.0001487names(model1)- 'coefficients'
- 'residuals'
- 'effects'
- 'rank'
- 'fitted.values'
- 'assign'
- 'qr'
- 'df.residual'
- 'xlevels'
- 'call'
- 'terms'
- 'model'
model1$fitted.values ##hat y
model1$coefficients- 1
- 8.16521739130434
- 2
- 18.6
- 3
- 21.2086956521739
- 4
- 18.6
- 5
- 18.6
- 6
- 29.0347826086957
- 7
- 13.3826086956522
- 8
- 23.8173913043478
- 9
- 13.3826086956522
- 10
- 21.2086956521739
- (Intercept)
- -2.2695652173913
- x
- 2.60869565217391
anova(model1) ## 회귀모형의 유의성 검정| Df | Sum Sq | Mean Sq | F value | Pr(>F) | |
|---|---|---|---|---|---|
| <int> | <dbl> | <dbl> | <dbl> | <dbl> | |
| x | 1 | 313.04348 | 313.043478 | 45.24034 | 0.0001486582 |
| Residuals | 8 | 55.35652 | 6.919565 | NA | NA |
a <- summary(model1)
ls(a)- 'adj.r.squared'
- 'aliased'
- 'call'
- 'coefficients'
- 'cov.unscaled'
- 'df'
- 'fstatistic'
- 'r.squared'
- 'residuals'
- 'sigma'
- 'terms'
summary(model1)$coef ## 회귀계수의 유의성 검정| Estimate | Std. Error | t value | Pr(>|t|) | |
|---|---|---|---|---|
| (Intercept) | -2.269565 | 3.212348 | -0.7065129 | 0.4999255886 |
| x | 2.608696 | 0.387847 | 6.7260939 | 0.0001486582 |
confint(model1, level = 0.95) ##회귀계수의 신뢰구간| 2.5 % | 97.5 % | |
|---|---|---|
| (Intercept) | -9.677252 | 5.138122 |
| x | 1.714319 | 3.503073 |
## beta +- t_alpha/2 (n-2) * se(beta)
qt(0.025, 8)
qt(0.975, 8)
-2.30600413520417
2.30600413520417
## y = beta1*x + epsilon
model2 <- lm(y ~ 0 + x, dt)
summary(model2)
Call:
lm(formula = y ~ 0 + x, data = dt)
Residuals:
Min 1Q Median 3Q Max
-4.0641 -1.5882 0.2638 1.4818 3.9359
Coefficients:
Estimate Std. Error t value Pr(>|t|)
x 2.3440 0.0976 24.02 1.8e-09 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 2.556 on 9 degrees of freedom
Multiple R-squared: 0.9846, Adjusted R-squared: 0.9829
F-statistic: 576.8 on 1 and 9 DF, p-value: 1.798e-09plot(y~x, data = dt,
xlab = "광고료",
ylab = "총판매액",
pch = 20,
cex = 2,
col = "darkorange")
abline(model1, col='steelblue', lwd=2)
abline(model2, col='green', lwd=2)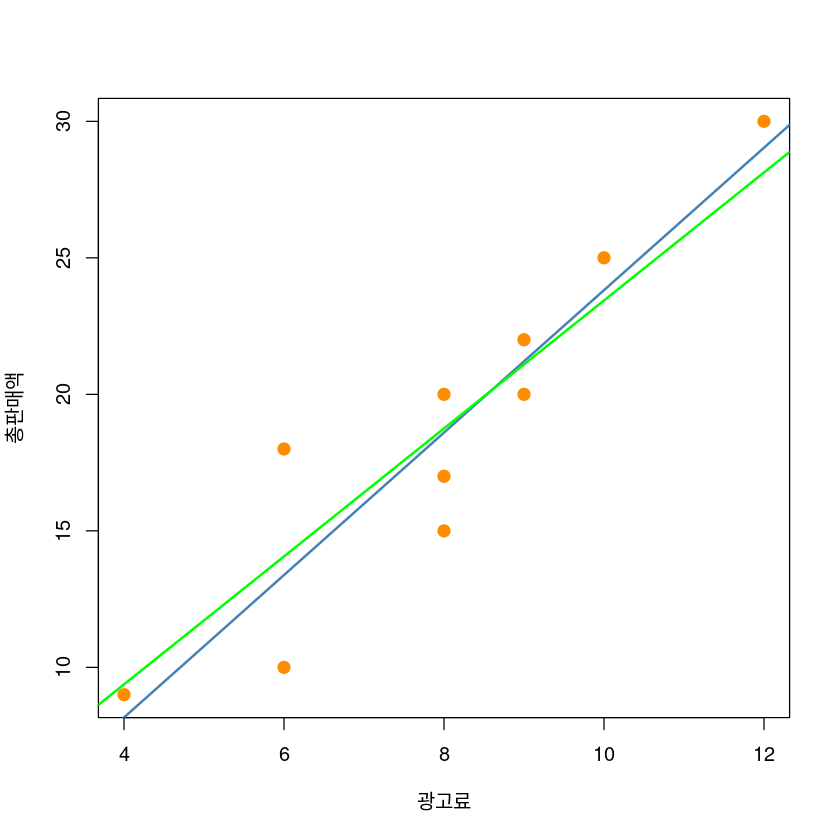
co <- coef(model1)ggplot(dt, aes(x, y)) +
geom_point(col='steelblue', lwd=1) +
geom_abline(intercept = co[1], slope = co[2], col='darkorange', lwd=1) +
xlab("광고료")+ylab("총판매액")+
theme_bw()+
theme(axis.title = element_text(size = 16))Warning message in geom_point(col = "steelblue", lwd = 1):
“Ignoring unknown parameters: `linewidth`”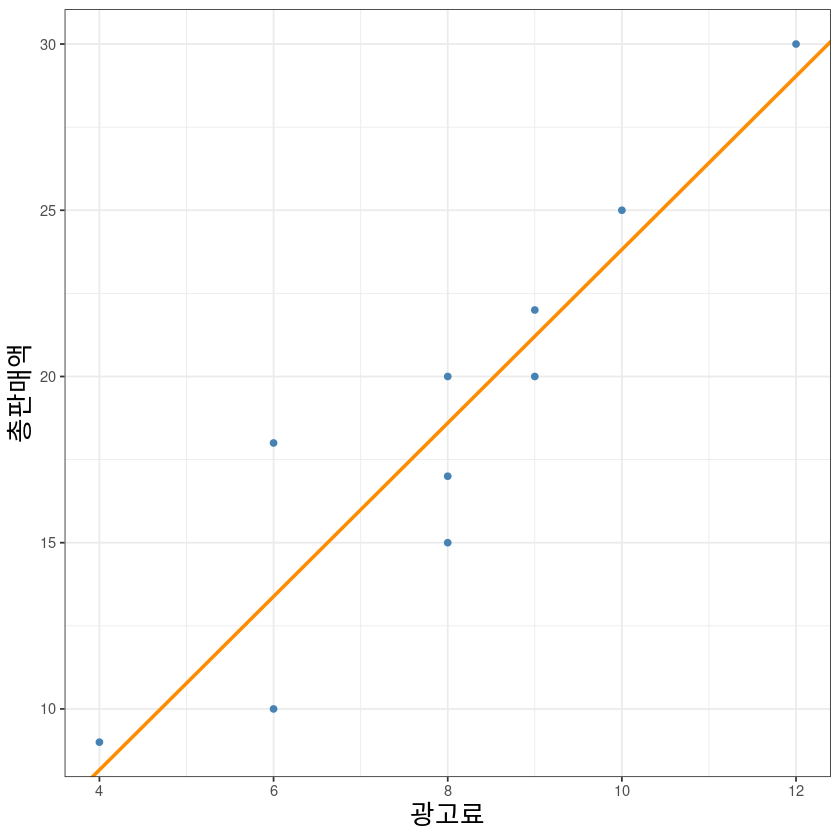
LSE 구하기
dt1 <- data.frame(
i = 1:nrow(dt),
x = dt$x,
y = dt$y,
x_barx = dt$x - mean(dt$x),
y_bary = dt$y - mean(dt$y))dt1$x_barx2 <- dt1$x_barx^2
dt1$y_bary2 <- dt1$y_bary^2
dt1$xy <-dt1$x_barx * dt1$y_barydt1| i | x | y | x_barx | y_bary | x_barx2 | y_bary2 | xy |
|---|---|---|---|---|---|---|---|
| <int> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> |
| 1 | 4 | 9 | -4 | -9.6 | 16 | 92.16 | 38.4 |
| 2 | 8 | 20 | 0 | 1.4 | 0 | 1.96 | 0.0 |
| 3 | 9 | 22 | 1 | 3.4 | 1 | 11.56 | 3.4 |
| 4 | 8 | 15 | 0 | -3.6 | 0 | 12.96 | 0.0 |
| 5 | 8 | 17 | 0 | -1.6 | 0 | 2.56 | 0.0 |
| 6 | 12 | 30 | 4 | 11.4 | 16 | 129.96 | 45.6 |
| 7 | 6 | 18 | -2 | -0.6 | 4 | 0.36 | 1.2 |
| 8 | 10 | 25 | 2 | 6.4 | 4 | 40.96 | 12.8 |
| 9 | 6 | 10 | -2 | -8.6 | 4 | 73.96 | 17.2 |
| 10 | 9 | 20 | 1 | 1.4 | 1 | 1.96 | 1.4 |
round(colSums(dt1),3)- i
- 55
- x
- 80
- y
- 186
- x_barx
- 0
- y_bary
- 0
- x_barx2
- 46
- y_bary2
- 368.4
- xy
- 120
### hat beta1 = S_xy / S_xx
##hat beta0 = bar y - hat beta_1 * bar x
beta1 <- as.numeric(colSums(dt1)[8]/colSums(dt1)[6])
beta0 <- mean(dt$y) - beta1 * mean(dt$x)cat("hat beta0 = ", beta0)hat beta0 = -2.269565cat("hat beta1 = ", beta1)hat beta1 = 2.608696평균반응, 개별 y 추정
############
## E(Y|x0), y = E(Y|x0) + epsilon
# x0 = 4.5
new_dt <- data.frame(x = 4.5)# hat y0 = hat beta0 + hat beta1 * 4.5
predict(model1,
newdata = new_dt,
interval = c("confidence"), level = 0.95)| fit | lwr | upr | |
|---|---|---|---|
| 1 | 9.469565 | 5.79826 | 13.14087 |
predict(model1, newdata = new_dt,
interval = c("prediction"), level = 0.95)| fit | lwr | upr | |
|---|---|---|---|
| 1 | 9.469565 | 2.379125 | 16.56001 |
dt_pred <- data.frame(
x = 1:12,
predict(model1,
newdata=data.frame(x=1:12),
interval="confidence", level = 0.95))
dt_pred| x | fit | lwr | upr | |
|---|---|---|---|---|
| <int> | <dbl> | <dbl> | <dbl> | |
| 1 | 1 | 0.3391304 | -6.2087835 | 6.887044 |
| 2 | 2 | 2.9478261 | -2.7509762 | 8.646628 |
| 3 | 3 | 5.5565217 | 0.6905854 | 10.422458 |
| 4 | 4 | 8.1652174 | 4.1058891 | 12.224546 |
| 5 | 5 | 10.7739130 | 7.4756140 | 14.072212 |
| 6 | 6 | 13.3826087 | 10.7597808 | 16.005437 |
| 7 | 7 | 15.9913043 | 13.8748223 | 18.107786 |
| 8 | 8 | 18.6000000 | 16.6817753 | 20.518225 |
| 9 | 9 | 21.2086957 | 19.0922136 | 23.325178 |
| 10 | 10 | 23.8173913 | 21.1945634 | 26.440219 |
| 11 | 11 | 26.4260870 | 23.1277880 | 29.724386 |
| 12 | 12 | 29.0347826 | 24.9754543 | 33.094111 |
dt_pred2 <- as.data.frame(predict(model1,
newdata=data.frame(x=1:12),
interval="prediction", level = 0.95))
dt_pred2| fit | lwr | upr | |
|---|---|---|---|
| <dbl> | <dbl> | <dbl> | |
| 1 | 0.3391304 | -8.5867330 | 9.264994 |
| 2 | 2.9478261 | -5.3751666 | 11.270819 |
| 3 | 5.5565217 | -2.2199297 | 13.332973 |
| 4 | 8.1652174 | 0.8663128 | 15.464122 |
| 5 | 10.7739130 | 3.8692308 | 17.678595 |
| 6 | 13.3826087 | 6.7738957 | 19.991322 |
| 7 | 15.9913043 | 9.5667143 | 22.415894 |
| 8 | 18.6000000 | 12.2379683 | 24.962032 |
| 9 | 21.2086957 | 14.7841056 | 27.633286 |
| 10 | 23.8173913 | 17.2086783 | 30.426104 |
| 11 | 26.4260870 | 19.5214047 | 33.330769 |
| 12 | 29.0347826 | 21.7358781 | 36.333687 |
names(dt_pred2)[2:3] <- c('plwr', 'pupr')dt_pred3 <- cbind.data.frame(dt_pred, dt_pred2[,2:3])barx <- mean(dt$x)
bary <- mean(dt$y)plot(y~x, data = dt,
xlab = "광고료",
ylab = "총판매액",
pch = 20,
cex = 2,
col = "grey",
ylim = c(min(dt_pred3$plwr), max(dt_pred3$pupr)))
abline(model1, lwd = 5, col = "darkorange")
lines(dt_pred3$x, dt_pred3$lwr, col = "dodgerblue", lwd = 3, lty = 2)
lines(dt_pred3$x, dt_pred3$upr, col = "dodgerblue", lwd = 3, lty = 2)
lines(dt_pred3$x, dt_pred3$plwr, col = "dodgerblue", lwd = 3, lty = 3)
lines(dt_pred3$x, dt_pred3$pupr, col = "dodgerblue", lwd = 3, lty = 3)
abline(h=bary,v=barx, lty=2, lwd=0.2, col='dark grey')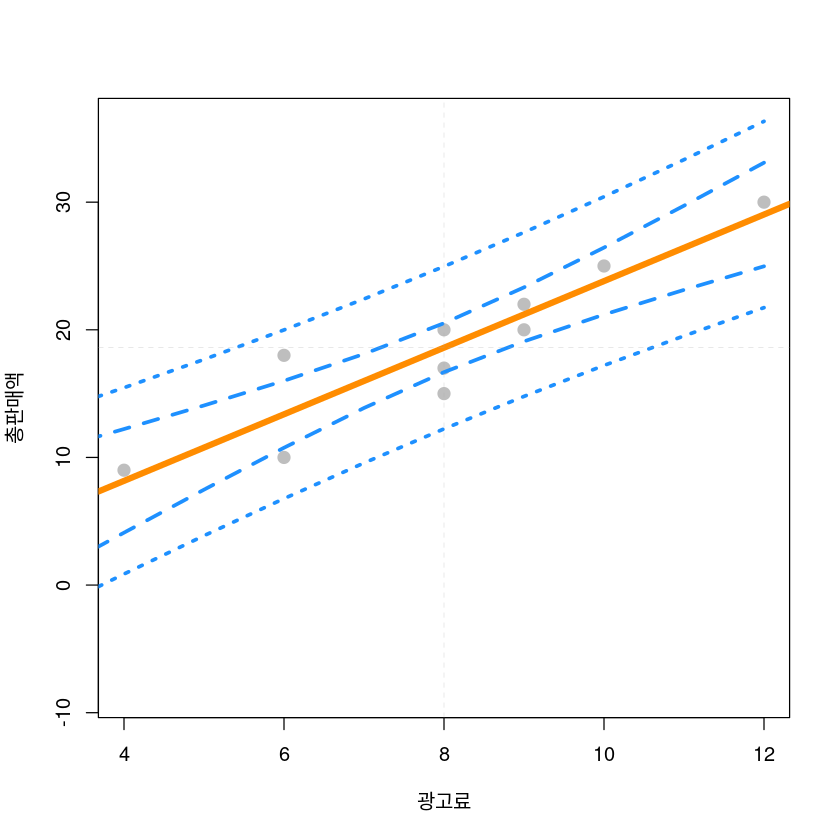
ggplot(dt_pred3, aes(x, fit)) +
geom_line(col='steelblue', lwd=2) +
xlab("")+ylab("")+
scale_x_continuous(breaks = seq(1,10))+
geom_line(aes(x, lwr), lty=2, lwd=1.5, col='darkorange') +
geom_line(aes(x, upr), lty=2, lwd=1.5, col='darkorange') +
geom_line(aes(x, plwr), lty=2, lwd=1.5, col='dodgerblue') +
geom_line(aes(x, pupr), lty=2, lwd=1.5, col='dodgerblue') +
geom_vline(xintercept = barx, lty=2, lwd=0.2, col='dark grey')+
geom_hline(yintercept = bary, lty=2, lwd=0.2, col='dark grey')+
theme_bw()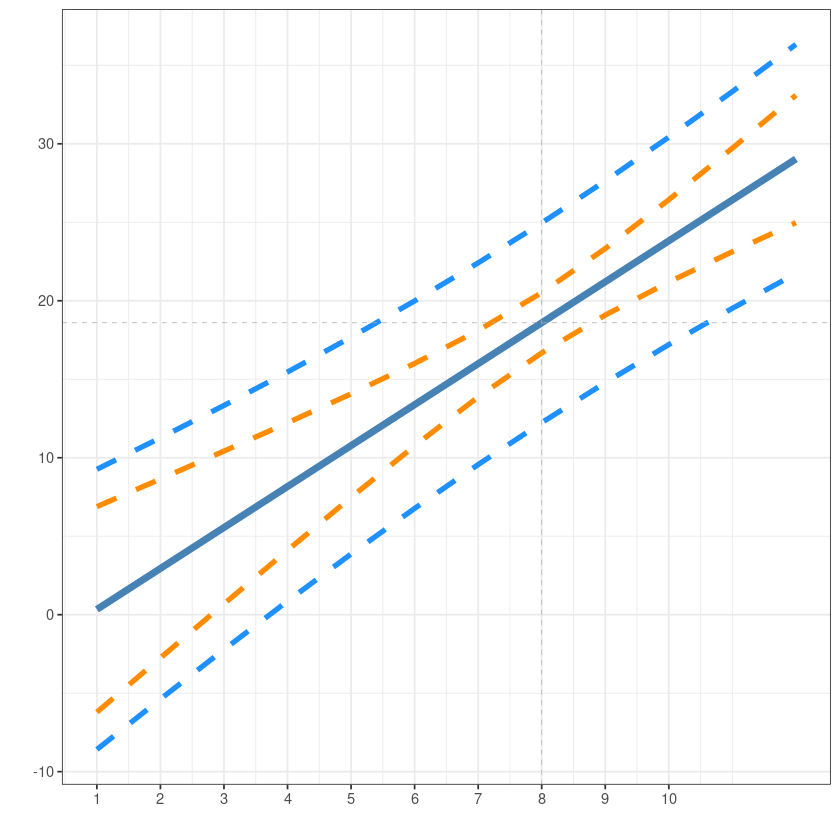
bb <- summary(model1)$sigma * ( 1 + 1/10 +(dt$x - 8)^2/46)
dt$ma95y <- model1$fitted + 2.306*bb
dt$mi95y <- model1$fitted - 2.306*bbggplot(dt, aes(x=x, y=y)) +
geom_point() +
geom_smooth(method=lm , color="red", fill="#69b3a2", se=TRUE) +
geom_line(aes(x, mi95y), col = 'darkgrey', lty=2) +
geom_line(aes(x, ma95y), col = 'darkgrey', lty=2) +
theme_bw() +
theme(axis.title = element_blank())`geom_smooth()` using formula = 'y ~ x'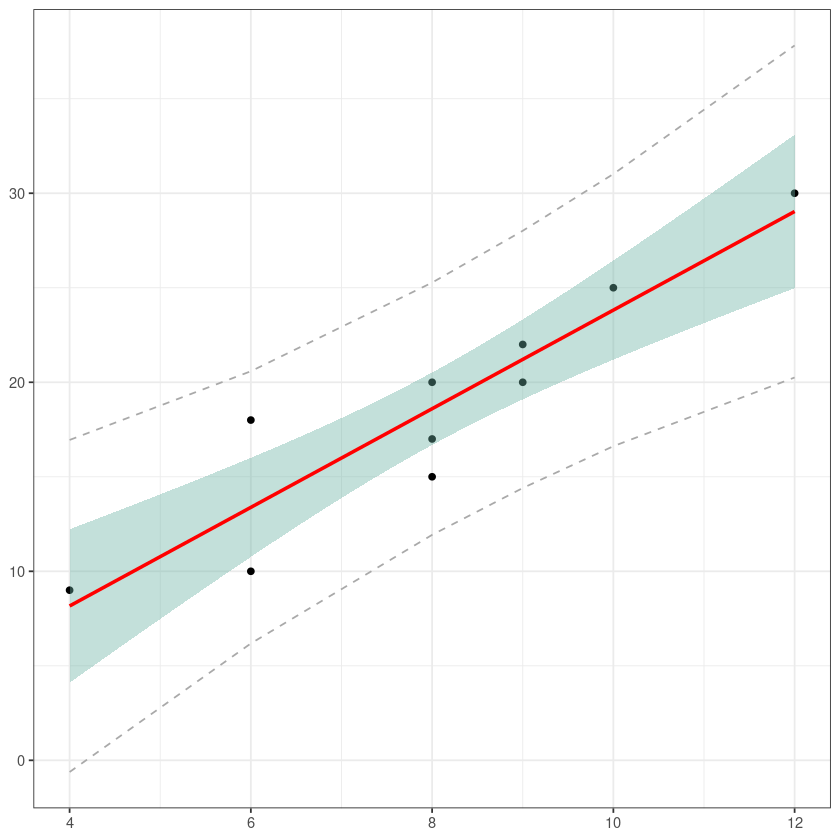
잔차분석
- epsilon : 선형성, 등분산성, 정규성, 독립성
dt| x | y | ma95y | mi95y |
|---|---|---|---|
| <dbl> | <dbl> | <dbl> | <dbl> |
| 4 | 9 | 16.94766 | -0.6172208 |
| 8 | 20 | 25.27254 | 11.9274568 |
| 9 | 22 | 28.01311 | 14.4042841 |
| 8 | 15 | 25.27254 | 11.9274568 |
| 8 | 17 | 25.27254 | 11.9274568 |
| 12 | 30 | 37.81722 | 20.2523444 |
| 6 | 18 | 20.58263 | 6.1825918 |
| 10 | 25 | 31.01741 | 16.6173744 |
| 6 | 10 | 20.58263 | 6.1825918 |
| 9 | 20 | 28.01311 | 14.4042841 |
dt$yhat <- model1$fitted# fitted.values(model1)
dt$resid <- model1$residuals
# resid(model1)
par(mfrow=c(1,2))
plot(resid ~ x, dt, pch=16, ylab = 'Residual')
abline(h=0, lty=2, col='grey')
plot(resid ~ yhat, dt, pch=16, ylab = 'Residual')
abline(h=0, lty=2, col='grey')
par(mfrow=c(1,1))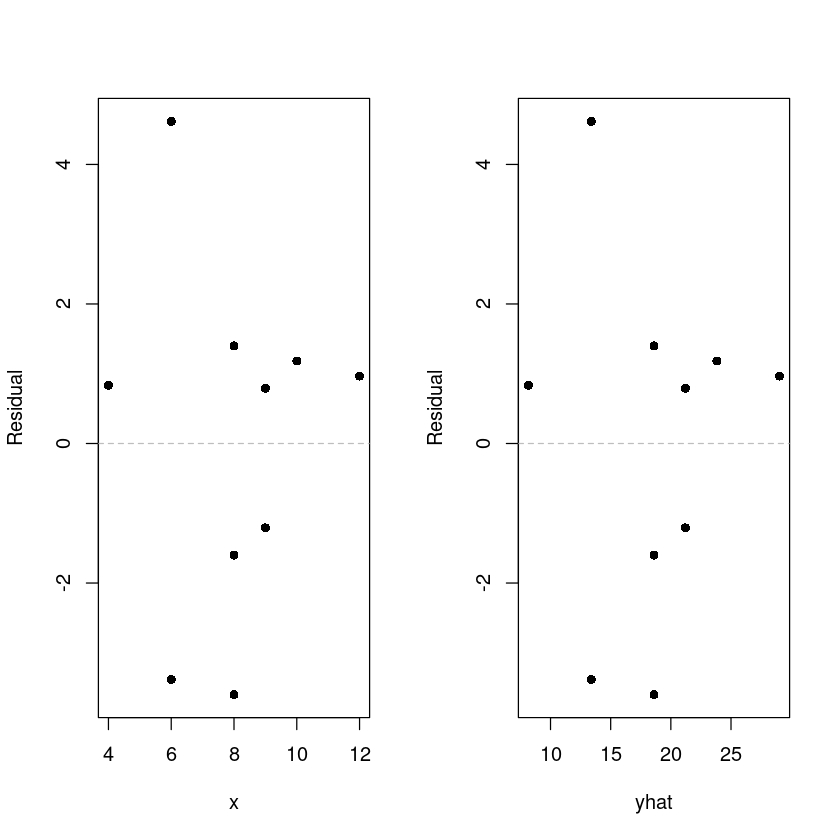
# 독립성검정 : DW test
library(lmtest)
##
dwtest(model1, alternative = "two.sided") #H0 : uncorrelated vs H1 : rho != 0
# dwtest(model1, alternative = "greater") #H0 : uncorrelated vs H1 : rho > 0
# dwtest(model1, alternative = "less")Loading required package: zoo
Attaching package: ‘zoo’
The following objects are masked from ‘package:base’:
as.Date, as.Date.numeric
Durbin-Watson test
data: model1
DW = 1.4679, p-value = 0.3916
alternative hypothesis: true autocorrelation is not 0## 정규분포 (QQ plot)
qqnorm(dt$resid, pch=16)
qqline(dt$resid, col = 2)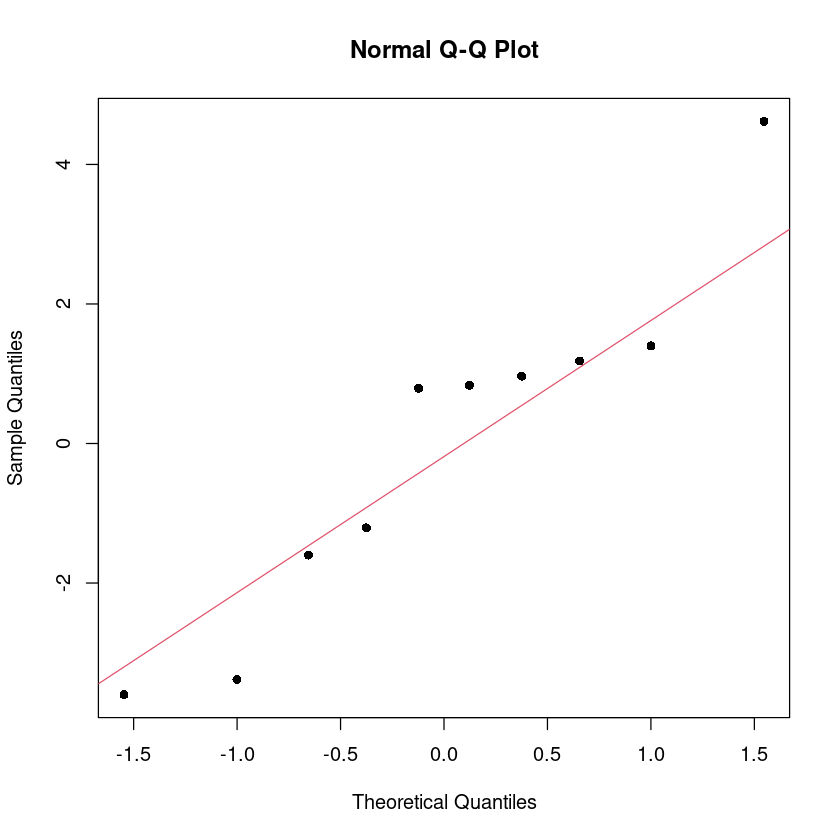
ggplot(dt, aes(sample = resid)) +
stat_qq() + stat_qq_line() +
theme_bw()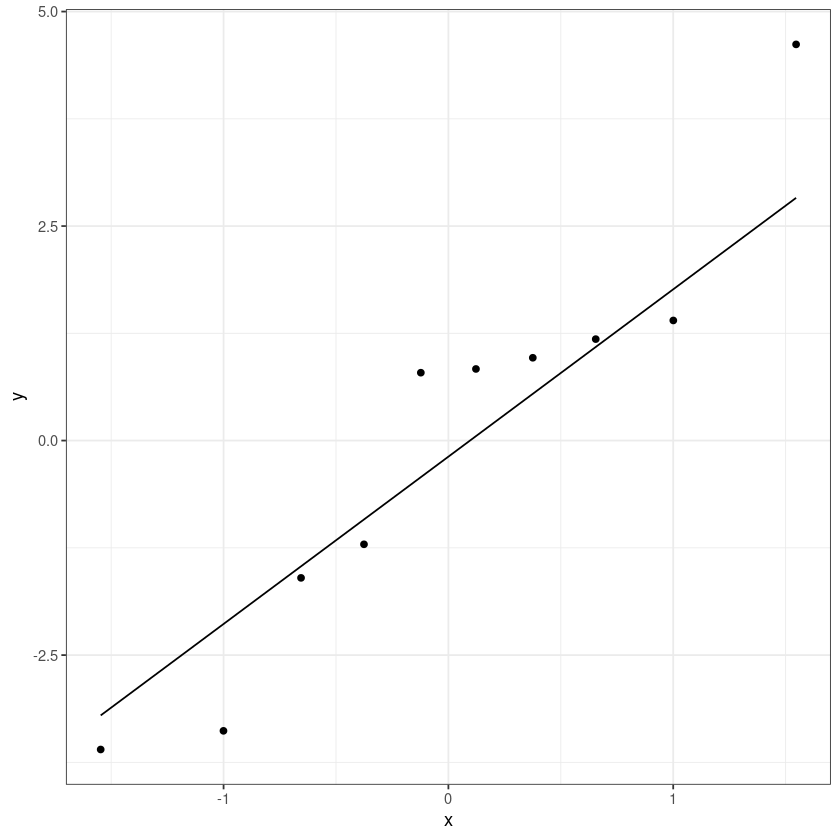
## 정규분포 검정
shapiro.test(dt$resid) ##shapiro-wilk test
#H0 : normal distributed vs H1 : not
Shapiro-Wilk normality test
data: dt$resid
W = 0.92426, p-value = 0.3939## 등분산성 검정
bptest(model1) #Breusch–Pagan test
# H0 : 등분산 vs H1 : 이분산
studentized Breusch-Pagan test
data: model1
BP = 1.6727, df = 1, p-value = 0.1959install.packages('UsingR')
library(UsingR)
data(father.son)Installing package into ‘/home/coco/R/x86_64-pc-linux-gnu-library/4.2’
(as ‘lib’ is unspecified)
also installing the dependency ‘HistData’
Loading required package: MASS
Loading required package: HistData
Loading required package: Hmisc
Loading required package: lattice
Loading required package: survival
Loading required package: Formula
Attaching package: ‘Hmisc’
The following objects are masked from ‘package:base’:
format.pval, units
Attaching package: ‘UsingR’
The following object is masked from ‘package:survival’:
cancer
names(father.son)- 'fheight'
- 'sheight'
lm.fit<-lm(sheight~fheight, data=father.son)
summary(lm.fit)
Call:
lm(formula = sheight ~ fheight, data = father.son)
Residuals:
Min 1Q Median 3Q Max
-8.8772 -1.5144 -0.0079 1.6285 8.9685
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 33.88660 1.83235 18.49 <2e-16 ***
fheight 0.51409 0.02705 19.01 <2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 2.437 on 1076 degrees of freedom
Multiple R-squared: 0.2513, Adjusted R-squared: 0.2506
F-statistic: 361.2 on 1 and 1076 DF, p-value: < 2.2e-16plot(sheight~fheight,
data=father.son,
pch=16, cex=0.5,
xlab="father’s height (inches)",
ylab="son’s height (inches)")
abline(lm.fit)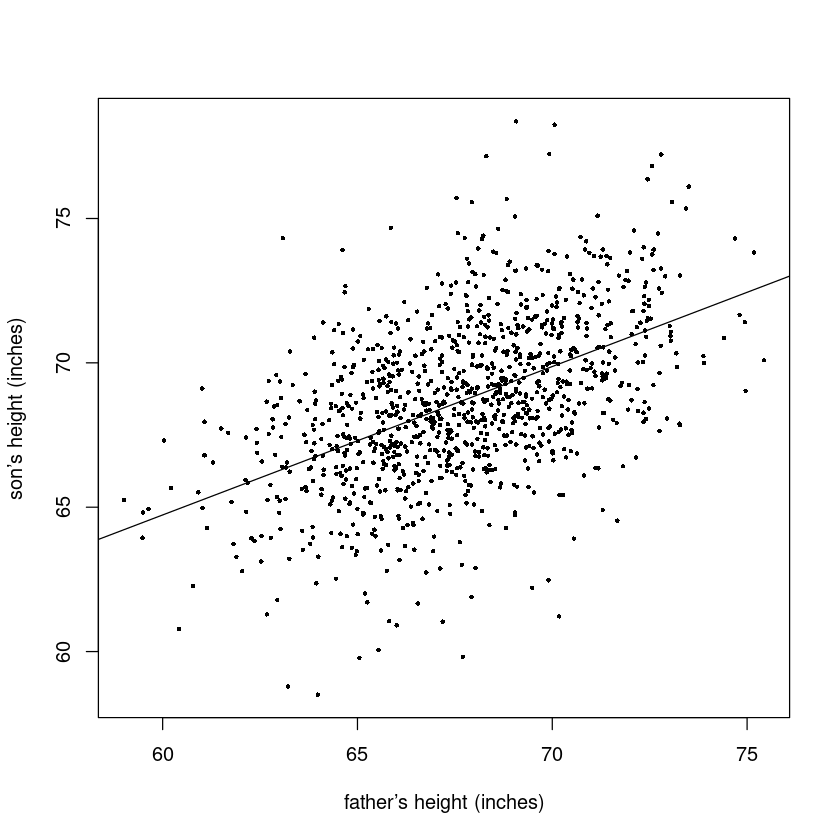
amazon<-read.csv("amazon.csv")
plot(High ~Year , amazon, pch=16)
lm.fit<-lm(High~Year, data=amazon)
summary(lm.fit)
confint(lm.fit)
par(mfrow=c(1,2))
scatter.smooth(x=1:length(amazon$Year), y=residuals(lm.fit), xlab="Year")
scatter.smooth(x=predict(lm.fit), y=residuals(lm.fit), xlab=expression(hat(y)))
library(lmtest)
dwtest(lm.fit)